Difference between revisions of "VolCT - Group 1A"
| Line 96: | Line 96: | ||
==== Comments on Reading Protocol ==== | ==== Comments on Reading Protocol ==== | ||
| − | # [[User:Npetrick|Npetrick]] 21: | + | # [[User:Npetrick|Npetrick]] 21:33, 18 February 2009 (UTC): Please provide your comments on the Reading protocol |
== 1A Questions to Group == | == 1A Questions to Group == | ||
Revision as of 21:33, 18 February 2009
Current Goals & Status:
- Goal: Benchmark intra/inter-reader variability for lung nodule volume measurements
- Status: Most of Dataset acquired; finalizing reader software
- Related Profile: Lung Vol Quantification - Measurement Activity
Work Documents
- Vol-CT - 1A Group Call Summaries
- Proposed 1A Protocol Outline (2008-11-13-v05)- For Comment from QIBA Members
- ---- Comments Bob Ford 12/16/2008
- RSNA Abstract SSK04-04: phantom study simialr to our proposal- For discussion on 12/18/2008 1A t-con.
- 2002-01-26: 1A Reader Study timeline for completion 1/1-4/15/2009- Please edit/comment
- 2009-02-02: Examples of Philips Medium and Detailed reconstruction filter slices- Please review/vote on appropriate filtering for 1A study
- 2009-02-02: 1A study 40 nodule slice date- 10mm ellipse -> 20 mm ellipse
Projects
May want to split these out to separate pages later
VolCT Lung Anthropomorphic Phantom Study
Objective:
Measure intra- & inter-reader bias and variability phantom lesions for:
- Uni-dimensional size measurement
- Semi-automatic 3D volumetric measure
May compare with fully automated algorithm(s)
Dataset:
Ground truth has been established by physical measurement "ex vivo“ on FDA phantom inserts.
Nodules (10 attached nodules)
- -10 & +100HU
- 10, 20 mm spheres
- 10 mm ovoid, lobulated, spiculated
Image Dataset
- 100 mAs exposures
- 0.75 & 5.0 mm slices
- 1 recon kernel
- Status:
- mostly acquired by FDA/CDRH/OSEL.
- Missing: 10 mm ovoid, spic, lob (Est: 12/01/08)
Acquisition Protocol:
- Scanner: Philips 16 slice
- Exposure (120 kVp): 100 mAs
- Slice thickness (50% overlap): 0.75 & 5.0 mm slices
- Recon kernel: Standard/medium (Still on table: Detail/Lung kernel)
- Pitch: 1.2
- 2 repeat scans
- 40 segmentations in set
The study is being conducted as a pilot. The size (data, readers) has not been selected for any specific level of significance.
Study Protocol:
Expert readers will measure/estimate nodule size from CT images.
Readers: 6 RadPharm radiologists
Software:
- Wendy will visit RadPharm and provide more info next week
- In-house review software (Siemens?)
- Semi-automated 3D volume software
- Uni-dimensional measure (RESIST)
- Which fully automated software?
Reading Session:
- Readers read all cases in 2 different reading sessions
- Random ordering
- One-dimensional measure
- Semi-automated segmentation
- Sessions separated by 3 week(?)
- Include duplicate cases within each read session (1/3-1/2 of cases for intra-reader estimates)
- Random ordering
- Time restrictions: Probably not (?)
- Specific instructions: Probably not (?)
- Readers read all cases in 2 different reading sessions
Analysis:
Estimate intra- and inter-reader variability in the different volume estimate
- Estimate bias from known truth
- Estimate variability
Compare the bias and variability with the different methods
Outstanding Issues:
1A Detailed Reading Protocol for Review
1A Protocol in Word
Comments on Reading Protocol
- Npetrick 21:33, 18 February 2009 (UTC): Please provide your comments on the Reading protocol
1A Questions to Group
Q1: 2009-02-02 Please select medium or detail reconstruction filter for 1A reader study
Philips Filter Descriptions
Detail: Medium-sharp filter, defined either for medium resolution images or moderate-noisy conditions of acquisitions (such as Posterior Fossa, chest and abdomen)
Medium: defined either to enhance the low contrast (for example Posterior Fossa) or to decrease the noise when the acquisition has been performed in relatively noisy conditions (such as large patient, thin slices and low mAs)
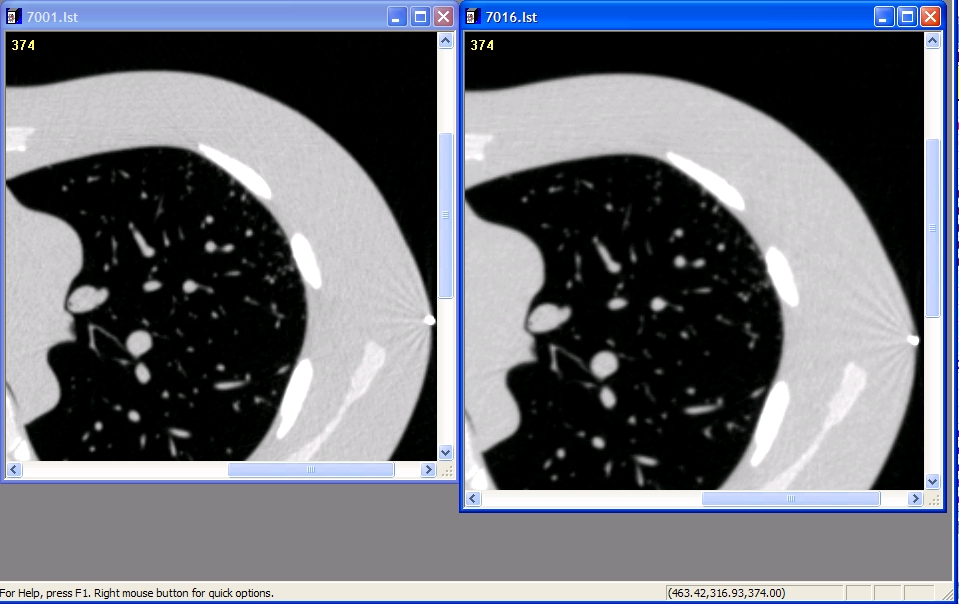
Powerpoint examples of nodules with Filters
Philips Medium/Detailed reconstruction filter slices.ppt
Example of Detailed (Left) and Medium (right) filter
Comments on filter type
- Npetrick 21:57, 6 February 2009 (UTC): My vote is for detailed since this is suggested by Philips for chest.
- Other votes?
- Other votes?
Nick - Are these the names that Philips uses? They used to use letters like "B" or "C". I think as long as the filter is not overenhancing, then this should be fine. I can't tell too much difference in the images, so differences look small (even the streaks look similar - overenhancing filters would change their appearance quite a bit). Mike
Q2: 2009-02-06 Please select output format for 1A reader study 3D segmentations
3D Data formats
Syntext of 3D Doctor Text Format:
Slice number
x1 y1
x2 y2
x3 y3
.
.
.
xn yn
x1 y1
END
Coordinates should be listed sequentially in polar-coordinate order
Syntext of PLY 3D Doctor Text Format (From Wikipedia):
PLY is a computer file format known as the Polygon File Format or the Stanford Triangle Format. It supports a relatively simple description of a single object as a list of nominally flat polygons. A variety of properties can be stored including: color and transparency, surface normals, texture coordinates and data confidence values. The format permits one to have different properties for the front and back of a polygon. There are two versions of the file format, one in ASCII, the other in binary.
PLY Wikipedia page
Example PLY
ply
format ascii 1.0
comment Mars model by Paul Bourke
element vertex 259200
property float x
property float y
property float z
element face 516960
property list uchar int vertex_indices
end_header
15081.5 -3.45644e+06 65.8061
15081 -3.45659e+06 197.422
15078.2 -3.45648e+06 329.009
15075.4 -3.45663e+06 460.597
15071.2 -3.4567e+06 592.148
15065.6 -3.45674e+06 723.653
15059.9 -3.457e+06 855.16
15050.7 -3.45674e+06 986.473
- Lots of vertices follow
14541.2 3.33642e+06 -698.464
14547.7 3.33663e+06 -571.58
14551.5 3.33649e+06 -444.589
14552.7 3.336e+06 -317.541
14556.9 3.33645e+06 -190.56
14558.7 3.33661e+06 -63.5247
3 0 721 1
3 721 0 720
3 1 722 2
3 722 1 721
3 2 723 3
3 723 2 722
- Lots of triangular facets follow
Comments on 3D data format
- Npetrick 21:57, 6 February 2009 (UTC): I'd vote for including both PLY and 3D Doctor formats if possible.
- David/Bob are both formats possible?
- Other 3D formats to consider? Segmentation and Markup Formats
- What is your vote?
I am not sure how to use the formats here very well, but I vote for being consistent with AIM (which I think uses a raster format). Mike